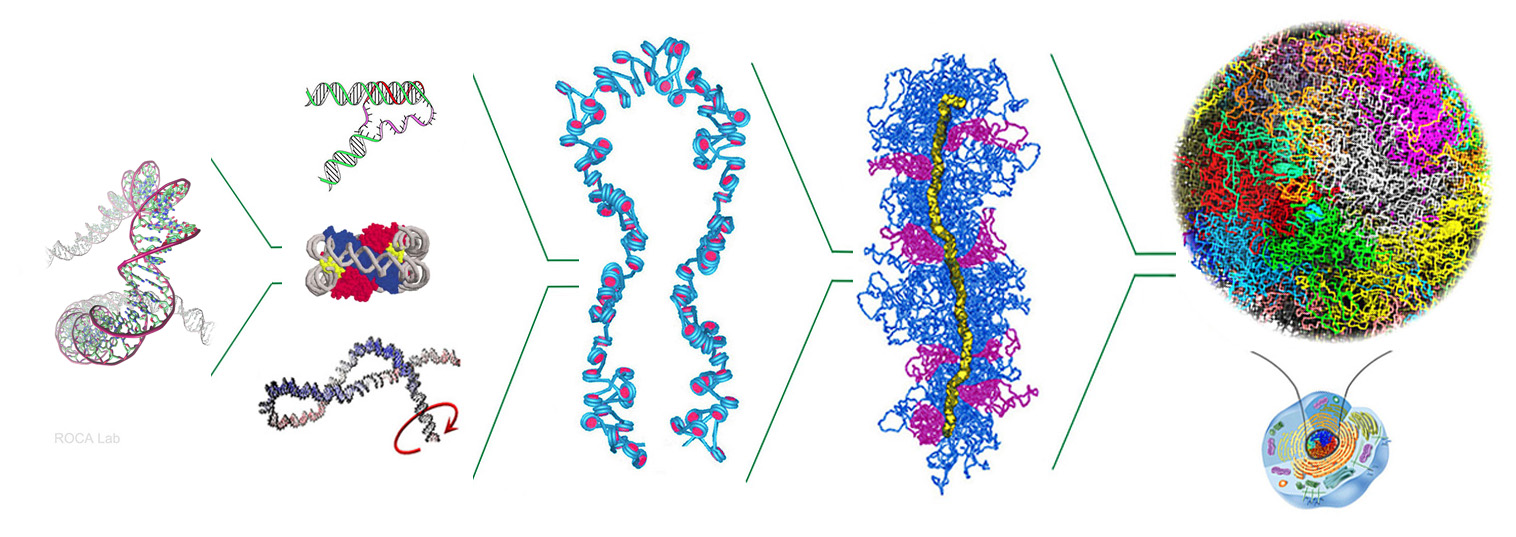
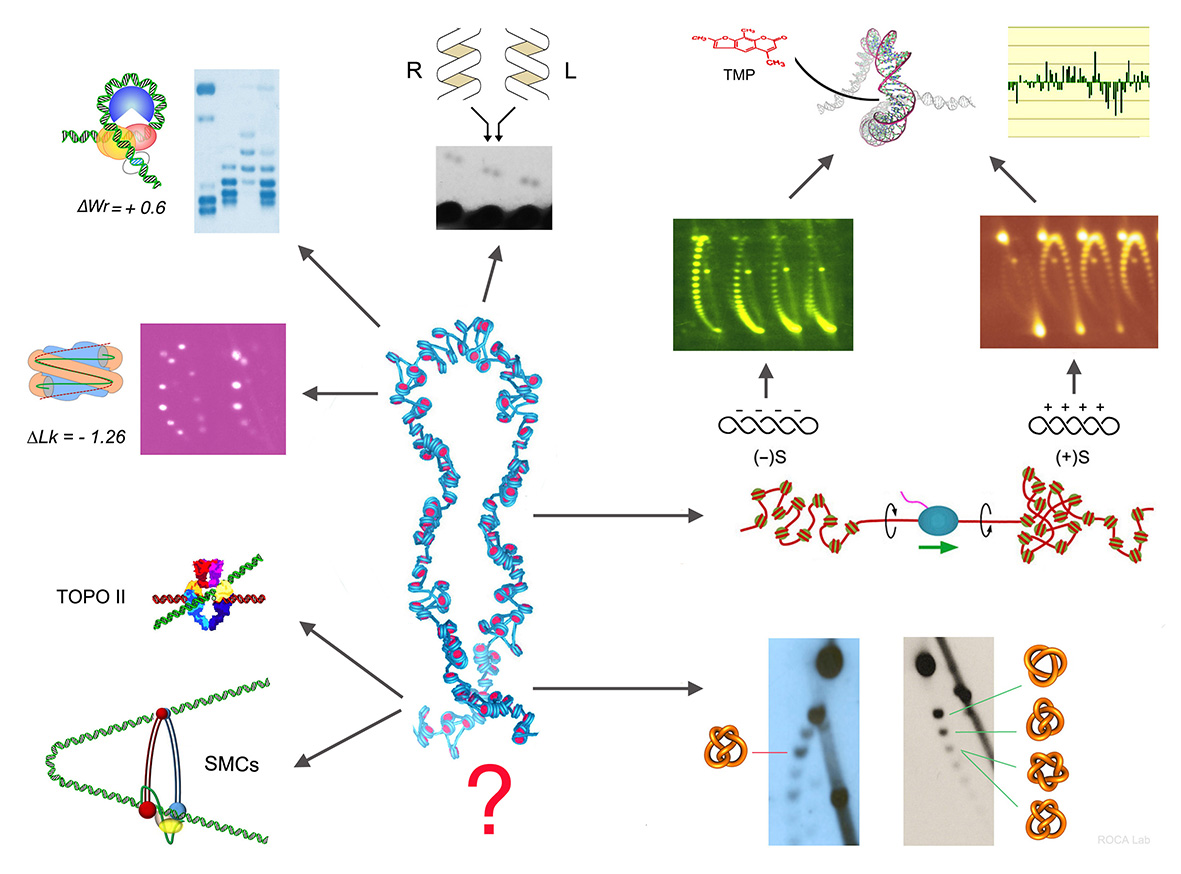
- Nucleosomal DNA has topological memory. Segura, Díaz-Ingelmo, Martínez-García, Ayats-Fraile, Nikolaou and Roca* bioRxiv DOI:10.1101/2023.05.21.541612. Nature Com -under review- (2024)
- Condensin pinches a short negatively supercoiled DNA loop during each round of ATP usage. Martínez-García, Dyson, Segura, Gutierrez-Escribano, Aragón, and Roca* EMBO J -e111913 (2023)
- Condensin minimizes topoisomerase II-mediated entanglements of DNA in vivo. Dyson, Segura, Martínez-García, Valdés, and Roca*. EMBO J - e105393 (2021)
- Transcriptional supercoiling boosts topoisomerase II-mediated knotting of intracellular DNA. Valdés, Coronel, Martínez-García, Segura, Dyson, Díaz-Ingelmo, Micheletti, and Roca* Nucleic Acids Res. 47:6946-6955 (2019)
- Quantitative disclosure of DNA knot chirality by high-resolution 2D-gel electrophoresis Valdés, Martínez-García, Segura, Dyson, Díaz-Ingelmo and Roca* Nucleic Acids Res. 47:e29 (2019)
- Intracellular nucleosomes constrain a DNA linking number difference of -1.26 that reconciles the Lk paradox. Segura, Joshi, Díaz-Ingelmo, Valdés, Dyson, Martínez-García and Roca* Nature Com 28:3989 (2018)
- DNA knots occur in intracellular chromatin. Valdes, Segura, Dyson,Martinez-Garcia and Roca* Nucleic Acids Res 46, 650-660 (2018)
- DNA Topology and Global Architecture of Point Centromeres. Diaz-Ingelmo, Martinez-Garcia, Segura, Valdes and Roca* Cell Reports 13, 667-677 (2015)
- Topoisomerase II minimizes DNA entanglements by proofreading DNA topology after DNA strand passage. Martinez-Garcia, Fernandez, Diaz-Ingelmo, Rodriguez-Campos, Manichanh and Roca* Nucleic Acids Res 42, 1821-1830 (2014)
- Chromatin regulates DNA torsional energy via topoisomerase II-mediated relaxation of positive supercoils. Fernandez, Diaz-Ingelmo, Martinez-Garcia and Roca* EMBO J 33, 1492-1501 (2014)
- Topoisomerase II regulates yeast genes with singular chromatin architectures. Nikolaou, Bermudez, Manichanh, Garcia-Martinez, Guigo, Perez-Ortin and Roca* Nucleic Acids Res. 41, 9243-9256 (2013)
- Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Joshi, Piña and Roca* Nucleic Acids Res 40, 7907-7915 (2012)
- Positional dependence of transcriptional inhibition by DNA torsional stress in yeast chromosomes. Joshi, Pina and Roca* EMBO J 29, 740-748 (2010)
- Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. Salceda, Fernandez and Roca* EMBO J 25, 2575-2583 (2006)
- DNA knots reveal a chiral organization of DNA in phage capsids. Arsuaga, Vazquez, McGuirk, Trigueros, Sumners and Roca* PNAS USA 102, 9165-9169 (2005)
- Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Arsuaga, Vazquez, Trigueros, Sumners and Roca* PNAS USA 99, 5373-5377 (2002)
- DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Roca, Berger, Harrison and Wang. PNAS USA 93, 4057-4062 (1996)
- DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Roca and Wang. Cell 77, 609-616 (1994).
- Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Roca, Ishida, Berger, Andoh, and Wang. PNAS USA 91, 1781-1785 (1994)
- The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Roca and Wang. Cell 71, 833-840 (1992)
- DNA topoisomerase II activity in nonreplicating, transcriptionally inactive, chicken late spermatids. Roca and Mezquita. EMBO J 8, 1855-1860 (1989).
|